Welcome to the Cram Lab!
Research Areas
The Cram Lab integrates observational, experimental, and computational modeling approaches to understand the distribution, dynamics, and diversity of all microbial species in different marine environments. For more information, contact Jacob Cram.
Ecology and Biogeochemistry of Marine Snow:
We focus primarily on observing and modeling microbial communities associated with sinking marine snow particles. Marine snow are sinking particles of primarily biological origin - they include aggregates of dead phytoplankton, fecal pellets from zooplankton, and other materials. They form primarily in surface waters and are consumed by zooplankton and bacteria. Marine snow and bacteria together are geologically significant because they drive a process called the “biological pump” . In this process, carbon dioxide is converted into planktonic biomass and eventually marine snow in the surface ocean. These particles sink into the deeper ocean where they are consumed by bacteria and converted back into carbon dioxide. In net, the biological pump is a process that uses energy (from sunlight) to transport carbon dioxide into the deep ocean. Bacteria, as the consumers of marine snow, are important regulators of this biological pump. The Cram Lab strives to observe and model dynamics of these particle associated bacteria. All of this work is done in collaboration with Fuchsman Lab, also at Horn Point.
Observations:
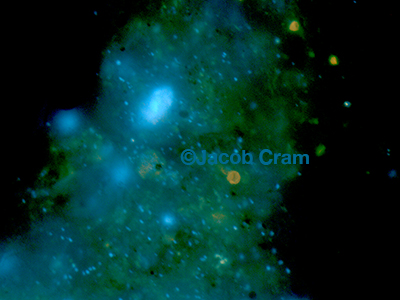
We strive to observe bacteria on particles in different regions of the ocean -- so far we are focused on two regions, the Eastern Tropical North Pacific (ETNP), and locally in the Chesapeake Bay and Mid-Atlantic Bight. Our ETNP work is done in collaboration with the Fuchsman lab at HPL and the Keil, Duval and Rocap Labs at University of Washington. We have been collecting particles with suspended sediment traps and studying the biology and chemistry of those particles. Our group has focused on examining bacterial concentrations on these particles using microscopy and molecular approaches.
In the Chesapeake Bay and Mid Atlantic Bight, we are collecting particles of different size fractions and trying to identify how many microorganisms are found on different kinds of particles and what kinds of microorganisms those are. We are also particularly interested in defining the rates that protists, bacteria and viruses emigrate off of or immigrate onto particles from the surrounding sea water and are developing methods to quantify this.
Modeling:
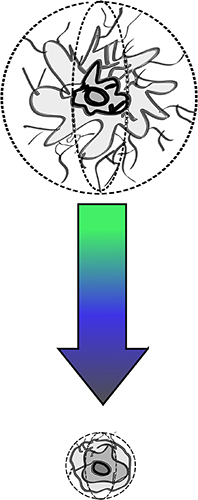
Dr. Cram’s previous work has involved modeling sinking particles to better understand how the environment and bacteria modulate the biological pump’s ability to transfer carbon to the deep ocean. We have been working on elaborating on this model, which considers particle size and dissolution dynamics, and allows scientists to specify the chemical composition of those particles. Ongoing work involves explicitly modeling the growth and death of bacteria and other microorganisms on particles and immigration of these organisms between particles and the surrounding sea-water.
Oyster Larval Microbiomes
Oyster aquaculture is a vital part of the economy of the state of Maryland and is a sustainable way to produce protein rich food. Understanding the health and physiology of larval oysters is an integral part improving oyster aquaculture in the Chesapeake bay and beyond. The bacteria that live in and on larval oysters can be referred to as their microbiome and this microbiome likely affects these larvae’s aforementioned health and physiology. In collaboration with the Plough and Gray labs we are investigating the microbiomes of larval oyster communities and exploring how microbiomes are dependent on larval strain (which is similar to a breed when we are talking about larger agricultural animals) and diet, and how strain and diet are associated with larval health and physiology. Towards this end, we are piggybacking on large oyster larval experiments and using molecular tools to observe the microbiome of larvae in different experimental groups and time points.
Opportunities
Interested in carrying out cutting edge research to learn about bacteria in marine environments? Come join us! Currently we have funding available for one graduate student and hope to be posting more opportunities here in the months ahead.
Graduate Students:
I would like to recruit, and have funding for, a graduate student for the Fall of 2019. If you are interested in joining my lab, please send me an email describing your research interests and a copy of your CV. All graduate students at Horn Point Laboratory are enrolled through the Marine Estuarine Environmental Sciences (MEES) Graduate Program. The application deadline for fall of 2019 is January 11.
While I commit to fully funding graduate students through their whole PhD (I won’t hire people if I can’t pay them) there are advantages to graduate students’ CV and the lab’s bottom line if we can net additional extramural (outside of the university) funding, such as the NSF Graduate Research Fellowship Program. I am enthusiastic about helping prospective graduate students apply for such extramural funding sources. Please contact me as soon as you are thinking about applying if interested.
Links
Publicly Available Data
Monthly measurements of microbial community structure, and associated environmental parameters from the San Pedro Ocean Time-Series.
Association network analysis
Cytoscape Tutorial that Cheryl Chow and I wrote for ecological association networks or whatever else you want to do with cytoscape.
LSA Tutorial. LSA is a useful way to look for time-lagged associations.
Here's the bitbucket page for extended local similarity analysis. It is maintained by my collaborator Li "Charlie" Xia
Code
Jacob Cram keeps most of his public code on bitbucket. His username is ohnoplus.
You can find there:
- One dimensional, size resolved, ballast and temperature dependent model of particle sinking
- Calculations of global variability in transfer efficency using the above model
- An example of how one can use Bray-Curtis similarity analysis to identify seasonal and long term patterns of variability in microbial community structure.
Some of J. Cram’s collaborators, Past and Present
- Mentors
- Curtis Deutsch - University of Washington (UW)
- Jed Fuhrman - University of Southern California (USC)
- Jim Kublin - Fred Hutchinson Cancer Research Center (FHCRC)
- Marine Microbial Ecology
- Clara Fuchsman - Horn Point Laboratory (HPL)
- David Needham - Monterey Bay Aquarium Research Institute
- Modeling
- Thomas Weber - Unviersity of Rochester
- Daniele Bianchi - University of California, Los Angles
- K. Allison Smith - Convoy
- Andrew McDonnell - University of Alaska Fairbanks
- Computational Biology
- Li Xia - Stanford University
- Fengzhu Sun - USC
- Andrew Fiore Gartland - FHCRC
- Oysters:
- Katie McFarland - HPL
- Louis Plough - HPL
- Matt Gray - HPL